Researchers have swapped the genome of gut germ E. coli for an artificial one
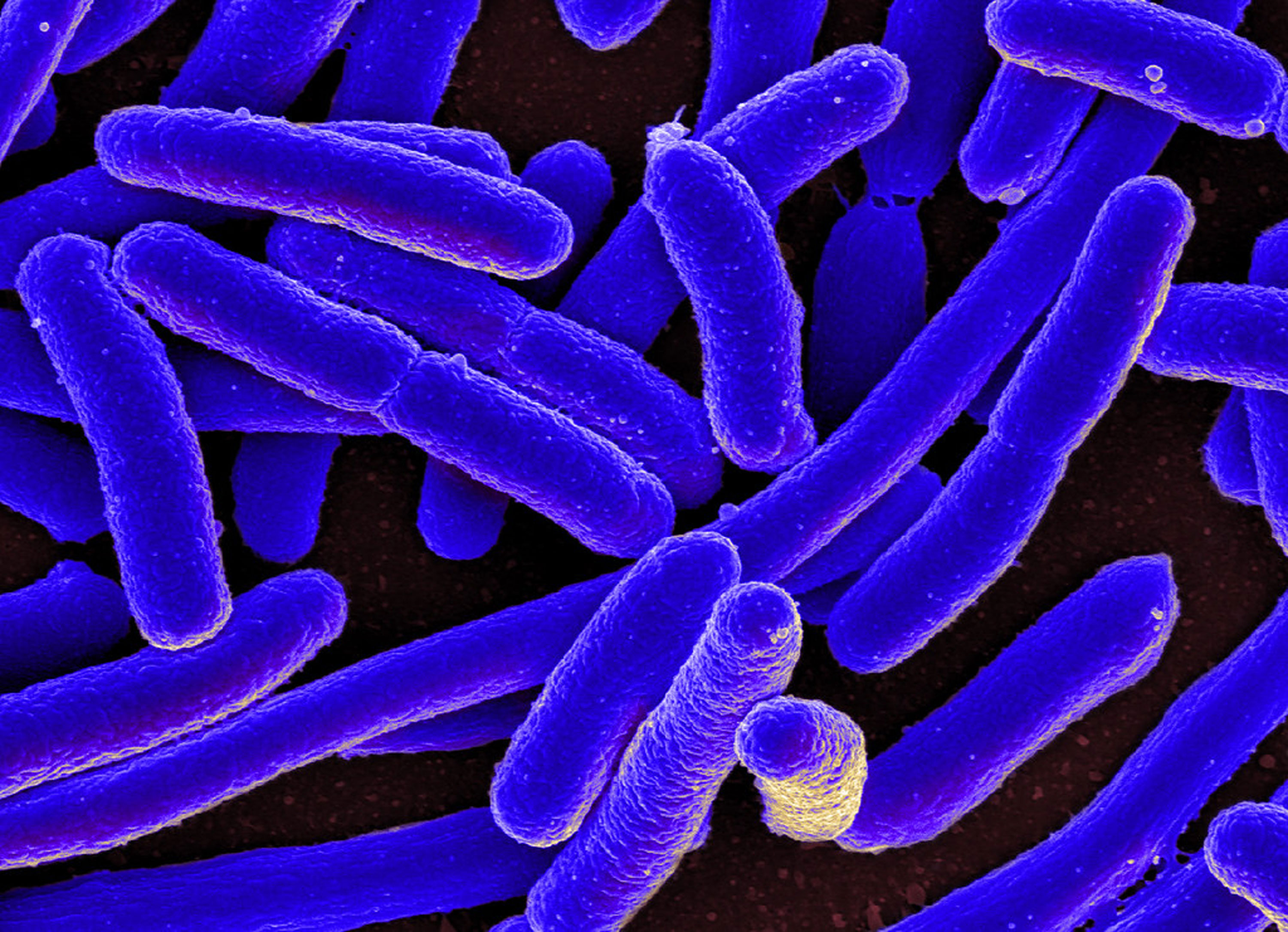
Researchers say they have replaced all the genes of E. coli bacteria with a complete copy of the genome synthesized in the lab. It’s a step toward creating germs that are genetically tailored to make a specific materials such as Kevlar or other polymers.
Scientists at the University of Cambridge report in Nature how, in a stepwise fashion, they gradually replaced the organism’s entire genome—it has 4 million DNA letters—with artificially made genes.
“It took two years, but we’d like to get this to the point where we can manufacture new synthetic genomes in less than a month,” says Jason Chin, a biologist with the UK Medical Research Council, who led the team. “That would massively accelerate the field, the number of things we can make and test.”
The first synthetic bacterial genomes were created in 2008 and 2010 at the J. Craig Venter Institute. But the E. coli genome, which is four times their size, sets a new record.
A separate consortium is trying to create baker’s yeast with artificial genes, but that project is not yet complete.
In replacing the bacterium’s genome, Chin’s team also simplified it by replacing some of the three-letter DNA instruction sets, or codons, that cells use to determine which of 20 amino acids they’ll add to a protein.
In the end, Chin’s E. coli has only 61 codons instead of the usual 64.
That means the new species of germs, called Syn61, don’t only have man-made genes, but also show that an organism can live with what the UK team calls a “compressed” genetic code.
“One is a technical achievement; the other tells you something fundamental about biology and how malleable the genetic code really is,” says Chin.
Simplifying the E. coli genome means the unused parts of the code are now free to do other things. For instance, they could be repurposed so that bacteria make proteins involving any of a couple of hundred amino acids that life doesn’t normally make use of. That could lead to the manufacture of unusual polymers in bacteria, like the material that goes into bulletproof vests.
There is also a scientific question, says Chin. Ever since the 1960s, when scientists first cracked the code, it’s been unclear exactly why it works the way it does—out of so many possibilities, why this one?
In 1968, Francis Crick, the co-discoverer of DNA’s chemical structure, proposed the “frozen accident” theory. Once basic life forms evolved, he suggested, the triplet codes got locked into place because any deviation from the universal program would be a big disadvantage. “By removing codons, we’re breaking that common language,” says Chin. “We’re unfreezing the code.”
Deep Dive
Biotechnology and health
How scientists traced a mysterious covid case back to six toilets
When wastewater surveillance turns into a hunt for a single infected individual, the ethics get tricky.
An AI-driven “factory of drugs” claims to have hit a big milestone
Insilico is part of a wave of companies betting on AI as the "next amazing revolution" in biology
The quest to legitimize longevity medicine
Longevity clinics offer a mix of services that largely cater to the wealthy. Now there’s a push to establish their work as a credible medical field.
There is a new most expensive drug in the world. Price tag: $4.25 million
But will the latest gene therapy suffer the curse of the costliest drug?
Stay connected
Get the latest updates from
MIT Technology Review
Discover special offers, top stories, upcoming events, and more.